(Part 4 of the saga of my escape from academia, see the original post that started everything, then also this and this.)
So in the end, an unexpected turn of luck. I am officially employed again. Out of academia. I work, in fact, in a mine.

Well, ok, not this kind of mine. Real miners are among the workers I respect more -doing a truly body-crushing job that is as fundamental to civilization as agriculture- but my skills as a coal miner would probably be more akin to those of Derek Zoolander:

Uhm. What I do is much more comfortable but somehow useful the same: text mining. Since last week I am (drum roll) a developer for Linguamatics, a Cambridge software company which develops the I2E text mining software.
What does it mean in practice? Everyone of my friends so far has asked me, so better now give a public answer. Now,
- I’m new to the whole business, so if some of my employers/colleagues is reading, please bear with me and point inaccuracies in the comments (I know the company’s CEO has landed on these pages)
- disclaimer: opinions/writings here are my own only and do not reflect those of my employers (and no, they didn’t ask me to write this).
I will make a simple example. Imagine you want to look a corpus of recipes, and you want to know what recipes involve blue cheese and pasta, but not meat (you happen to be a vegetarian). Now, unless your recipe corpus is explicitly structured this way, you have little chance of finding what you want by, say, “googling” it. You can’t look for cheese, pasta and meat, because a simple search engine has no hope of knowing that Stilton is a cheese and that lasagne contain meat.

Text mining solves this kind of basic problem, because it includes knowledge on the items you’re looking for. You can have a structured vocabolary (in jargon this is called usually an ontology) that looks like this:
Dairy
Cheese
Fresh Cheese
Feta
...
Matured Cheese
Cheddar
Wensleydale
...
Blue Cheese
Stilton
Gorgonzola
So when the program sees “Stilton” it knows that it belongs to the category of “dairy”, which in turns includes “cheese”, all down to “blue cheese”. While instead if the software sees “Wensleydale cheese with blue berries”, it won’t match it, even if it contains “blue” and “cheese”, because it doesn’t belong to the category of blue cheeses. So now instead of looking for words, like you do on Google or Bing, you look for concepts -which is much more similar to what humans do.
Now, this already would be useful, but it’s only the beginning of the story. The real power comes when you want to look at relationships between things. To understand this, let’s make another example.
Suppose you have an ecosystem of animals living together. You want to know, for example, which birds are killers of which mammal (silly example probably, but I’m not an ecologist!). Now, an ontology like the one above would only solve part of the problem, because of course you can build one to let the program know that “tiger” is a mammal and “hawk” is a bird. But imagine just looking for mammals and birds occurring together in sentences in books and articles. We can have sentences like:
- “Hawks hunt rabbits”
- “Vultures eat wildebeest corpses”
- “Pheasants are hunted by tigers”
Here you need also to understand language (and we enter a daunting field known as natural language processing). What you want is only sentence #1. Sentence #2 is not what you want, even if there is “BIRD eat MAMMAL”, because a wildebeest corpse is not an alive wildebeest. The third is actually the opposite relationship, but it would match a simple text search of “BIRD hunt MAMMAL”.
And we’re not even entering the problem of context: if you want to know what tigers do in woods, and look on a simple search engine, chances are that you’ll find a lot of information about this guy instead than about big cats hiding in the jungle. Again, we have to understand what we’re reading, not simply looking in bags of words.

That is what makes text mining so useful and complex: it doesn’t mean simply to look for documents. Text mining tools basically read the text for you and sort-of understand it, extracting the information you need within a billion of other informations. That’s why it is called mining: it’s like recovering the few diamonds from tons of rocks.
And to imagine how many rocks you have, well, let’s look at PubMed, the database of scientific papers’ abstracts. PubMed indexes a bit less than 21 million articles, and something like 500.000 articles are added each year. Now, imagine you want to gauge what the scientific community has found about some particular correlation nobody ever thought of condensing in a review article, but which you know is in the scientific literature. Text mining helps (among other things) to solve exactly that.
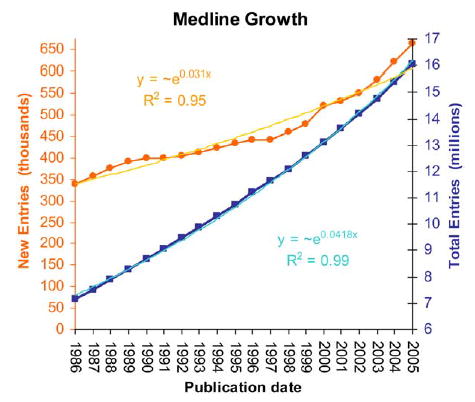
So what a former molecular biologist/biophysicist does there? Well, the above should make it more clear. Biomedical scientific literature is at the same time one of the most valuable, largest, fastest growing and most complex corpuses of information. Finding relevant information there is essential. And of course to develop the tools that do that in the best way possible you need also people who know what it’s all about -and there (among with others) I am.
About the differences between academia and a company job, on one hand there would be lot to write, and on the other hand it’s still too soon to put it all down. But most importantly, this is just about me. While instead we have to go deeper into the academia situation for everyone, and I have ideas to begin to do that.
Well, good luck.
I was just reading your *most cited* work (“Goodbye Academia”), when you just uploaded this new post.
My experience resembles the “second path” you described when decided to let the world know you are leaving academia. I am still there, 41 years old, few months in Berlin and then back in Italy as, again, 1 (one) year contract reseacher.
My approach was led by the idea that things can be changed, and I am (was) involved in the non-staff (“precari”) movement within my University (Siena), with the belief that consciousness and knowledge of our state may improve the situation, at least to give us a new “life plan” based on clear perspectives.
Hardly.
But still I persist on that.
You are now trying your new life plan; I would say in due time. Later would be too late.
just wondering about metaphors and this tool. they can be tricky, uh? it’s a fascinating field, even if, after my thesis on computer analysis of psychotherapy transcription, it completely lost its grip on me…